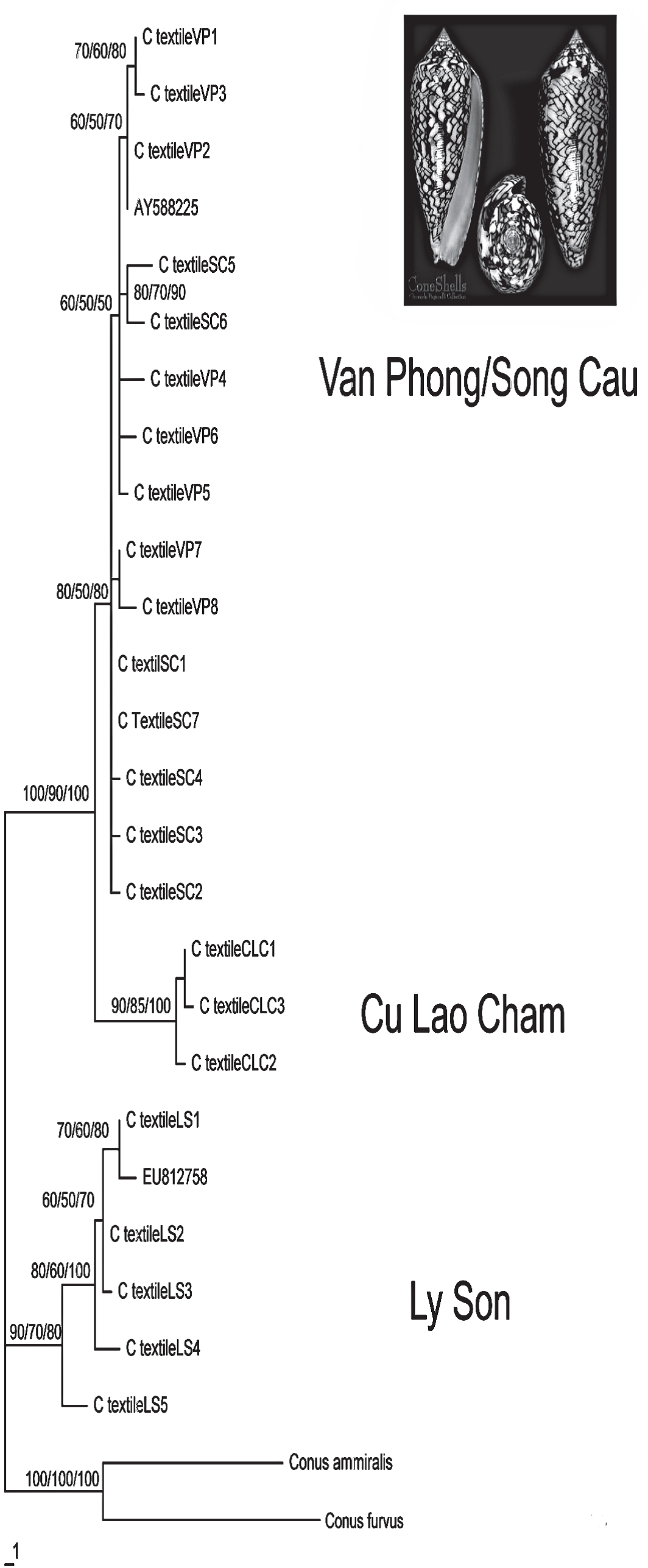
 To investigate the population genetics of Conus textile, we sequenced the COI mtDNA gene of 45 individuals from four different localities of the Southern Central Coast of Vietnam (Ly Son, Cu Lao Cham, Song Cau and Van Phong). C. textile populations exhibited high genetic variation with 23 haplotypesdiscovered. The common haplotype found occurring in three populations, was not observed at Cu Lao Cham.The sequence differences vary from 0.2 – 1% and 0.5 – 4.8% within and between populations. Ly Son and Cu Lao Cham populations have genetically separated on the phylogram. Meanwhile the haplotypes of Van Phong and Song Cau populations have been mixed. As only a low level of gene flow observed, populations of C. textile at Southern Central Vietnam are suggested to be recently derived.
To investigate the population genetics of Conus textile, we sequenced the COI mtDNA gene of 45 individuals from four different localities of the Southern Central Coast of Vietnam (Ly Son, Cu Lao Cham, Song Cau and Van Phong). C. textile populations exhibited high genetic variation with 23 haplotypesdiscovered. The common haplotype found occurring in three populations, was not observed at Cu Lao Cham.The sequence differences vary from 0.2 – 1% and 0.5 – 4.8% within and between populations. Ly Son and Cu Lao Cham populations have genetically separated on the phylogram. Meanwhile the haplotypes of Van Phong and Song Cau populations have been mixed. As only a low level of gene flow observed, populations of C. textile at Southern Central Vietnam are suggested to be recently derived.
The population genetics of Conus textile Linnaeus, 1758 from the Southern central coast of Vietnam
Related
- Phylogeny of zoonotic parasites in fresh and brackish water fish in Vietnam (21-7-2014 12:12:31)
- Isolation, identification of luminous Vibrio in diseased cultured common seahorse (Hippocampus kuda) (04-8-2014 02:03:24)
- Phylogenetic relationship of giant clams (Tridacna spp.) collected in the South and Central coast of Vietnam (04-8-2014 02:01:42)
- Feeding mode of cone snail (Conus spp.) and phylogenetic relationships (04-8-2014 01:59:46)
Others
- About us (17-7-2014 09:35:40)
- SVM30022GR0249 "Management and restoration mangrove forests in the north of Khanh Hoa province - Toward sustainable landscape" (25-10-2023 09:44:51)
- VINIF.2022.DA00021 "DNA metabarcoding and integrated information database - Implication for Ichthyoplankton survey and fisheries management in Vietnam" (25-10-2023 09:53:54)
- Riverscape Genetics to Inform Natural History of Exploited Fishes in the Lower Mekong River Basin (23-10-2017 11:43:14)
